COMPANY INSIGHT
Sponsored by ImmuMap
Personalized Medicine: Improved Accuracy of Neoepitope Prediction by Real Life Testing of T Cell Receptor Recognition using Novel High Throughput Technology
Personalized cancer vaccination is mainstay but hampered by inadequate performance of T cell neoepitope predictors and low capacity methods for pre-existent T cell responses. ImmuMap offers next-generation identification of 1031 neoepitopes in parallel using dCODETM. Our mission is to advance the breadth of knowledge achievable per volume of patient sample.
Immuno-oncology (IC), has developed much more rapidly than any other mode of treatment in oncology for the past decade. This breakthrough last year led to the nobel prize in Physiology or Medicine being awarded to the pioneering work of James Alisson and Tasuku Honjo.
The three main classes of drugs in IC count, (i) the checkpoint inhibitor blockers and other antibody-based strategies, (ii) the chimeric antigen receptor (CAR)- or T cell receptor (TCR)-transduced T cells, and (iii) neoantigen targeting strategies. The checkpoint inhibitor blockers were first to hit the market and followed in 2017 with approval of some CAR T cell therapies. Recent efforts have now gravitated towards developing neoantigen therapeutic strategies. Here, T cell recognizing “new” epitopes occurring due to mutations in the protein sequence are targeted and expanded, a strategy which has also shown clinical promise.
We aim to optimize the neoantigen selection strategy as depicted in figure 1. The process shared by most companies for neoantigen selection is similar: extracted tumor cells are tested by next-generation sequencing (NGS) for mutations as compared to autologous healthy tissue. Based on this information potential neoepitopes are predicted using a ranking algorithm according to epitope/major histocompatibility complex (MHC) binding affinity predictions, antigen expression and other parameters. Before and during treatment blood samples are drawn for immune monitoring. These data are used to feed the prediction algorithm and over time increase the validity of the output. It is important to realize, though, that the obtained data from such immune monitoring become skewed as only immune cells recognizing the selected targets are investigated.
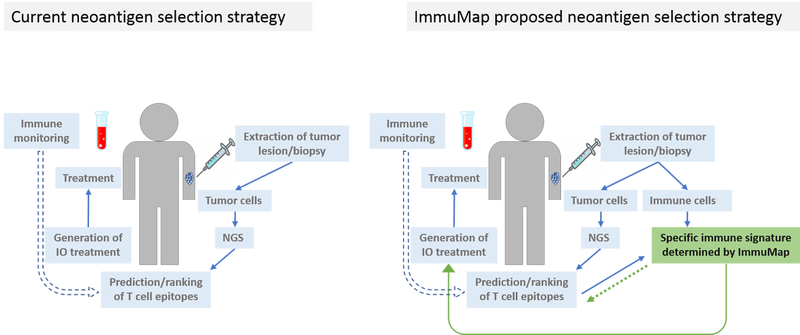
The neoantigen players differ in their treatment strategy post target selection and by the proprietary in silico predictor they develop. Only seldomly, however, are the selected targets tested in a cellular system beforehand of starting patient treatment. Numerous companies operate within the hot neoantigen field, and according to the Immune-Oncology Research Trends website (May 23 2019), the most promising of these are Advaxis, CureVac, Moderna and BioNtech. The first three companies select neoantigens using bioinformatics only, while BioNtech previously utilized a mouse breast cancer model. They have also expressed interest in a similar method to the dCODE™ conducted in the lab of professor Sine Reker Hadrup, a co-founder of ImmuMap.
It has been estimated (Saini et al., Annals of Oncology 2017) that on average only 4.7% of the selected targets used in neoantigen published trials led to detectable populations of tumor-recognizing T cells, so the room for improvement is large.
The reason for this lack of screening for pre-existent T cell responses is probably the scarcity of immune cells present in a tumor lesion, and at the same time high cell numbers are required for individual testing of T cell epitopes using standard methods. If we assume the number of tumor-infiltrating lymphocytes which can be isolated from a tumor lesion is approximately 2 million, this will only be sufficient for screening for T cells recognizing 2 or 10 epitopes using standard ELISpot (enzyme-linked immunospot) or MHC multimer staining for flow cytometry, respectively.
It is well established that multi-target strategies are advantageous in immune therapy of cancer and often as many as 20 personalized candidate epitopes are used for treatment. We here introduce the unique dCODETM platform, which enables us to deconvolute for as many as 1031 different T cell populations in the same amount of material. We have the means to directly test and rank multiple in silico predicted neoepitopes and additionally include also predicted non-epitope peptides. Data from such studies can thus be used for designing a truly personalized cancer vaccine in addition to optimizing the prediction algorithm.
It is important to stress that the currently used “personalized” treatment is built of patient-specific NGS analysis on one side and the outcome of prediction algorithms trained on multiple non-personalized data on the other.
Table 1. Head-to-head comparison of central elements in our proposed neoantigen selection strategy and the common one.
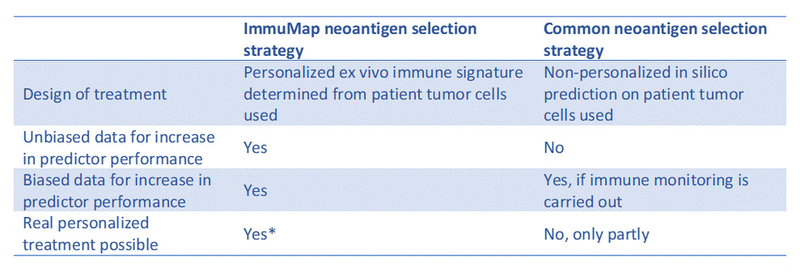
*Requires fast-tracking of reagent production, ImmuMap analysis and NGS provider analysis
We propose that a pre-treatment testing of a broad selection of candidate neoepitopes will return the necessary data for designing superior neoantigen treatment regimens with the resulting better clinical outcome.
